github/ropensci/phylogram
We use CiteLang to clone this repository, and parse found dependency files to generate this credit table and badge. For each research software repository, we assign a credit split (0.5 or 50/50) to say that the research software gets 50% of the credit, and the remaining 50% is shared by the dependencies. The same holds true for the children of dependencies up until we reach a minimum level of credit in the tree, at which point we stop parsing.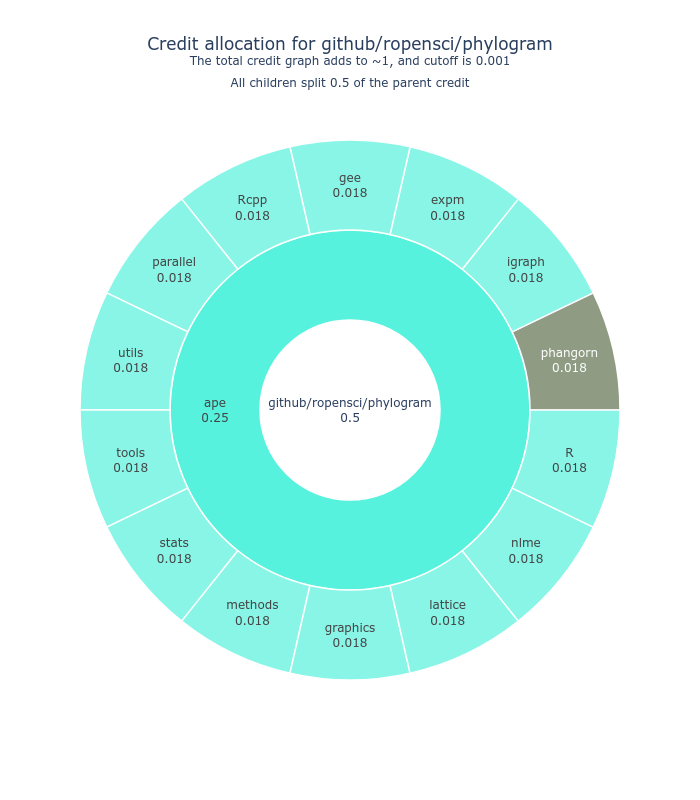
Software Credit
| Manager | Name | Credit |
|---|---|---|
| cran | stats | 0.127 |
| cran | utils | 0.104 |
| cran | methods | 0.102 |
| cran | graphics | 0.092 |
| cran | R | 0.089 |
| cran | parallel | 0.073 |
| cran | tools | 0.07 |
| cran | MASS | 0.05 |
| cran | Matrix | 0.021 |
| cran | ape | 0.017 |
| cran | tinytest | 0.015 |
| cran | Rmpfr | 0.014 |
| cran | sfsmisc | 0.014 |
| cran | RColorBrewer | 0.014 |
| cran | grDevices | 0.014 |
| cran | pkgKitten | 0.012 |
| cran | rbenchmark | 0.012 |
| cran | inline | 0.012 |
| DESCRIPTION | github/ropensci/phylogram | 0.01 |
| cran | lattice | 0.009 |
| cran | SASmixed | 0.009 |
| cran | Hmisc | 0.009 |
| cran | chron | 0.007 |
| cran | latticeExtra | 0.007 |
| cran | KernSmooth | 0.007 |
| cran | grid | 0.007 |
| cran | rgl | 0.007 |
| cran | digest | 0.004 |
| cran | withr | 0.004 |
| cran | testthat | 0.004 |
| cran | tcltk | 0.004 |
| cran | stats4 | 0.004 |
| cran | scales | 0.004 |
| cran | igraphdata | 0.004 |
| cran | graph | 0.004 |
| cran | pkgconfig | 0.004 |
| cran | magrittr | 0.004 |
| cran | igraph | 0.004 |
| cran | Rcpp | 0.004 |
| cran | xtable | 0.003 |
| cran | seqLogo | 0.003 |
| cran | seqinr | 0.003 |
| cran | rmarkdown | 0.003 |
| cran | prettydoc | 0.003 |
| cran | magick | 0.003 |
| cran | knitr | 0.003 |
| cran | Biostrings | 0.003 |
| cran | quadprog | 0.003 |
| cran | fastmatch | 0.003 |
| cran | phangorn | 0.001 |
| cran | expm | 0.001 |
| cran | gee | 0.001 |
| cran | nlme | 0.001 |
Note that credit values are rounded and expanded (so shared dependencies are represented as one record) and may not add to 1.0. Rounded values that hit zero are removed.
- Generated by CiteLang