github/nanoporetech/medaka
We use CiteLang to clone this repository, and parse found dependency files to generate this credit table and badge. For each research software repository, we assign a credit split (0.5 or 50/50) to say that the research software gets 50% of the credit, and the remaining 50% is shared by the dependencies. The same holds true for the children of dependencies up until we reach a minimum level of credit in the tree, at which point we stop parsing.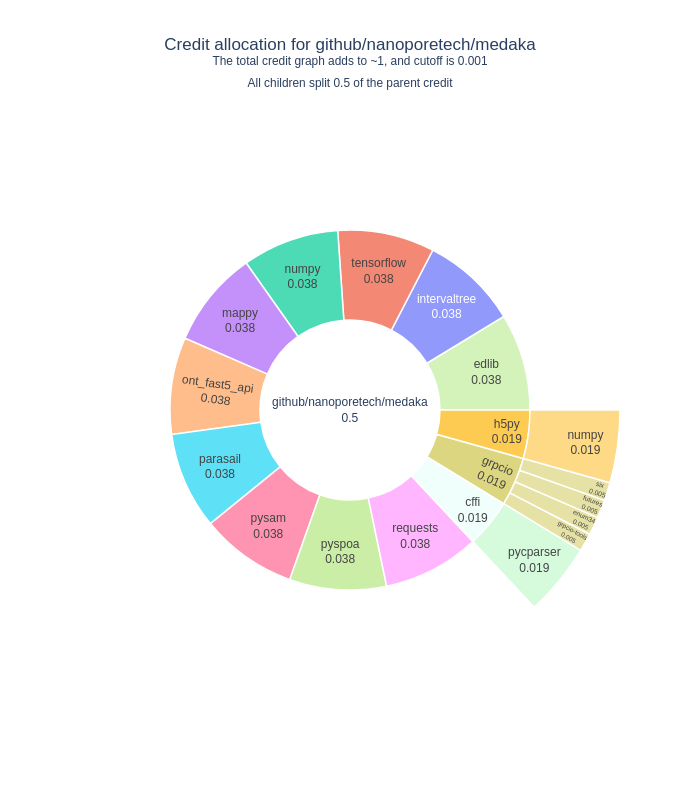
Software Credit
| Manager | Name | Credit |
|---|---|---|
| pypi | numpy | 0.249 |
| pypi | edlib | 0.076 |
| pypi | intervaltree | 0.076 |
| pypi | mappy | 0.076 |
| pypi | pysam | 0.076 |
| pypi | pyspoa | 0.076 |
| pypi | pycparser | 0.075 |
| pypi | h5py | 0.023 |
| pypi | six | 0.022 |
| pypi | packaging | 0.022 |
| pypi | enum34 | 0.019 |
| pypi | futures | 0.019 |
| pypi | progressbar33 | 0.019 |
| pypi | win-inet-pton | 0.011 |
| pypi | PySocks | 0.011 |
| pypi | charset-normalizer | 0.011 |
| pypi | idna | 0.011 |
| pypi | chardet | 0.011 |
| pypi | certifi | 0.011 |
| pypi | urllib3 | 0.011 |
| pypi | grpcio | 0.01 |
| requirements.txt | github/nanoporetech/medaka | 0.01 |
| pypi | setuptools | 0.01 |
| pypi | protobuf | 0.01 |
| pypi | keras | 0.003 |
| pypi | tensorflow-estimator | 0.003 |
| pypi | tensorboard | 0.003 |
| pypi | tensorflow-io-gcs-filesystem | 0.003 |
| pypi | wrapt | 0.003 |
| pypi | typing-extensions | 0.003 |
| pypi | termcolor | 0.003 |
| pypi | opt-einsum | 0.003 |
| pypi | libclang | 0.003 |
| pypi | keras-preprocessing | 0.003 |
| pypi | google-pasta | 0.003 |
| pypi | gast | 0.003 |
| pypi | flatbuffers | 0.003 |
| pypi | astunparse | 0.003 |
| pypi | absl-py | 0.003 |
| pypi | cffi | 0.001 |
| pypi | tensorflow | 0.001 |
| pypi | ont_fast5_api | 0.001 |
| pypi | parasail | 0.001 |
| pypi | requests | 0.001 |
Note that credit values are rounded and expanded (so shared dependencies are represented as one record) and may not add to 1.0. Rounded values that hit zero are removed.
- Generated by CiteLang