github/institut-de-genomique/HAPO-G
We use CiteLang to clone this repository, and parse found dependency files to generate this credit table and badge. For each research software repository, we assign a credit split (0.5 or 50/50) to say that the research software gets 50% of the credit, and the remaining 50% is shared by the dependencies. The same holds true for the children of dependencies up until we reach a minimum level of credit in the tree, at which point we stop parsing.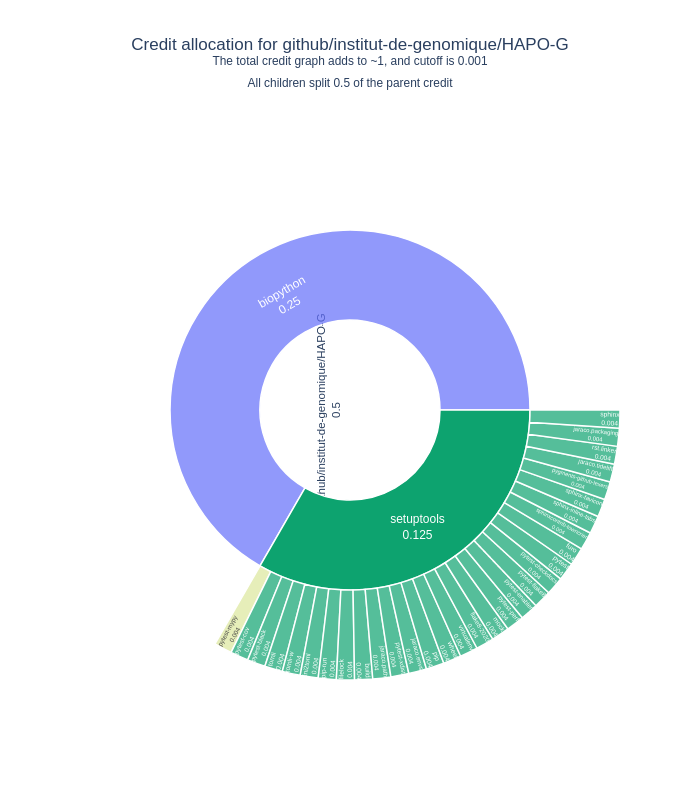
Software Credit
| Manager | Name | Credit |
|---|---|---|
| pypi | biopython | 0.495 |
| pypi | pytest | 0.023 |
| pypi | filelock | 0.02 |
| pypi | pytest-xdist | 0.018 |
| pypi | virtualenv | 0.018 |
| pypi | tomli | 0.017 |
| pypi | tomli-w | 0.017 |
| pypi | pytest-black | 0.016 |
| pypi | pip-run | 0.016 |
| pypi | build | 0.016 |
| pypi | jaraco.path | 0.016 |
| pypi | jaraco.envs | 0.016 |
| pypi | pip | 0.016 |
| pypi | wheel | 0.016 |
| pypi | flake8-2020 | 0.016 |
| pypi | mock | 0.016 |
| pypi | pytest-perf | 0.016 |
| pypi | pytest-enabler | 0.016 |
| pypi | pytest-flake8 | 0.016 |
| pypi | pytest-checkdocs | 0.016 |
| pypi | furo | 0.016 |
| pypi | sphinxcontrib-towncrier | 0.016 |
| pypi | sphinx-inline-tabs | 0.016 |
| pypi | sphinx-favicon | 0.016 |
| pypi | pygments-github-lexers | 0.016 |
| pypi | jaraco.tidelift | 0.016 |
| pypi | rst.linker | 0.016 |
| pypi | jaraco.packaging | 0.016 |
| pypi | sphinx | 0.016 |
| requirements.txt | github/institut-de-genomique/HAPO-G | 0.01 |
| pypi | setuptools | 0.006 |
| pypi | mypy | 0.004 |
| pypi | attrs | 0.004 |
| pypi | six | 0.002 |
| pypi | process-tests | 0.002 |
| pypi | hunter | 0.002 |
| pypi | fields | 0.002 |
| pypi | coverage | 0.002 |
| pypi | pytest-cov | 0.001 |
| pypi | typing-extensions | 0.001 |
| pypi | validate-pyproject | 0.001 |
| pypi | importlib-metadata | 0.001 |
| pypi | pyproject-fmt | 0.001 |
| pypi | tomlkit | 0.001 |
| pypi | configupdater | 0.001 |
| pypi | packaging | 0.001 |
Note that credit values are rounded and expanded (so shared dependencies are represented as one record) and may not add to 1.0. Rounded values that hit zero are removed.
- Generated by CiteLang