github/bioexcel/biobb_wf_flexdyn
We use CiteLang to clone this repository, and parse found dependency files to generate this credit table and badge. For each research software repository, we assign a credit split (0.5 or 50/50) to say that the research software gets 50% of the credit, and the remaining 50% is shared by the dependencies. The same holds true for the children of dependencies up until we reach a minimum level of credit in the tree, at which point we stop parsing.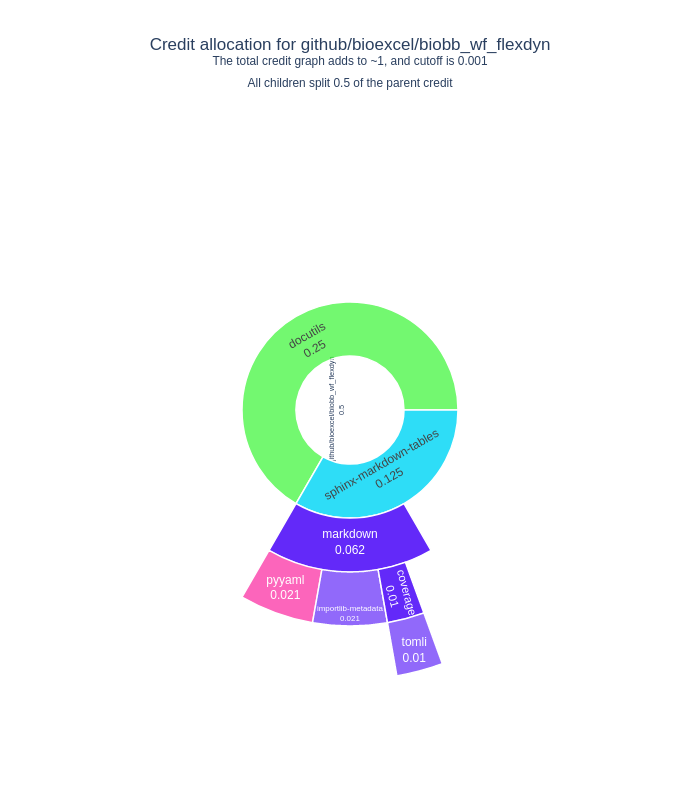
Software Credit
| Manager | Name | Credit |
|---|---|---|
| pypi | docutils | 0.495 |
| pypi | pyyaml | 0.162 |
| pypi | tomli | 0.16 |
| requirements.txt | github/bioexcel/biobb_wf_flexdyn | 0.01 |
| pypi | importlib-resources | 0.007 |
| pypi | pytest-flake8 | 0.007 |
| pypi | pytest-mypy | 0.007 |
| pypi | pytest-black | 0.007 |
| pypi | pytest-perf | 0.007 |
| pypi | flufl.flake8 | 0.007 |
| pypi | pyfakefs | 0.007 |
| pypi | packaging | 0.007 |
| pypi | pytest-enabler | 0.007 |
| pypi | pytest-cov | 0.007 |
| pypi | flake8 | 0.007 |
| pypi | pytest-checkdocs | 0.007 |
| pypi | pytest | 0.007 |
| pypi | ipython | 0.007 |
| pypi | jaraco.tidelift | 0.007 |
| pypi | sphinx-lint | 0.007 |
| pypi | furo | 0.007 |
| pypi | rst.linker | 0.007 |
| pypi | jaraco.packaging | 0.007 |
| pypi | sphinx | 0.007 |
| pypi | typing-extensions | 0.007 |
| pypi | zipp | 0.007 |
| pypi | sphinx-markdown-tables | 0.005 |
| pypi | markdown | 0.005 |
| pypi | coverage | 0.002 |
| pypi | importlib-metadata | 0.002 |
Note that credit values are rounded and expanded (so shared dependencies are represented as one record) and may not add to 1.0. Rounded values that hit zero are removed.
- Generated by CiteLang