github/LSARP/ProteomicsQC
We use CiteLang to clone this repository, and parse found dependency files to generate this credit table and badge. For each research software repository, we assign a credit split (0.5 or 50/50) to say that the research software gets 50% of the credit, and the remaining 50% is shared by the dependencies. The same holds true for the children of dependencies up until we reach a minimum level of credit in the tree, at which point we stop parsing.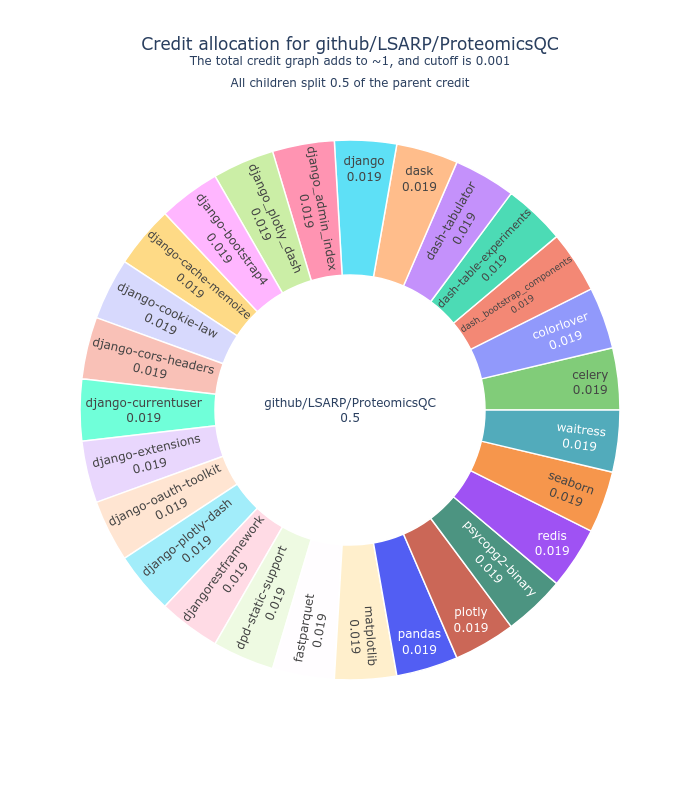
Software Credit
| Manager | Name | Credit |
|---|---|---|
| pypi | celery | 0.037 |
| pypi | colorlover | 0.037 |
| pypi | dash_bootstrap_components | 0.037 |
| pypi | dash-table-experiments | 0.037 |
| pypi | dash-tabulator | 0.037 |
| pypi | dask | 0.037 |
| pypi | django | 0.037 |
| pypi | django_admin_index | 0.037 |
| pypi | django_plotly_dash | 0.037 |
| pypi | django-bootstrap4 | 0.037 |
| pypi | django-cache-memoize | 0.037 |
| pypi | django-cookie-law | 0.037 |
| pypi | django-cors-headers | 0.037 |
| pypi | django-currentuser | 0.037 |
| pypi | django-extensions | 0.037 |
| pypi | django-oauth-toolkit | 0.037 |
| pypi | django-plotly-dash | 0.037 |
| pypi | djangorestframework | 0.037 |
| pypi | dpd-static-support | 0.037 |
| pypi | fastparquet | 0.037 |
| pypi | matplotlib | 0.037 |
| pypi | pandas | 0.037 |
| pypi | plotly | 0.037 |
| pypi | psycopg2-binary | 0.037 |
| pypi | redis | 0.037 |
| pypi | seaborn | 0.037 |
| pypi | waitress | 0.037 |
| requirements.txt | github/LSARP/ProteomicsQC | 0.01 |
Note that credit values are rounded and expanded (so shared dependencies are represented as one record) and may not add to 1.0. Rounded values that hit zero are removed.
- Generated by CiteLang