medrxivr
If you’ve ever done some kind of literature analysis, you’ve probably also done some web scraping to retrieve content, or otherwise struggled with ancient APIs to download some subset of data. If you are particularly interested in preprints for medicine, then you might have heard of MedrXiv, a database of preprints for health sciences. Forget about scraping this server, a newly published tool from ROpenSci called medrxivr is all you need to download preprint data.

Are you already familiar with this software? We encourage you to contribute to the research software encyclopedia and annotate the respository:
otherwise, keep reading!
- What is medrxivr?
- How do I cite it?
- How do I get started?
- How do I contribute to the software survey
- Where can I learn more?
What is medrxivr?
MedrXxiv is a database of pre-prints for health sciences, and MedrXivr is the “archiver” for that database. You can install it
install.packages("devtools")
devtools::install_github("ropensci/medrxivr")
library(medrxivr)
and easily download your own copy of the database:
preprint_data <- mx_api_content() # from live API
preprint_data <- mx_snapshot() # snapshot
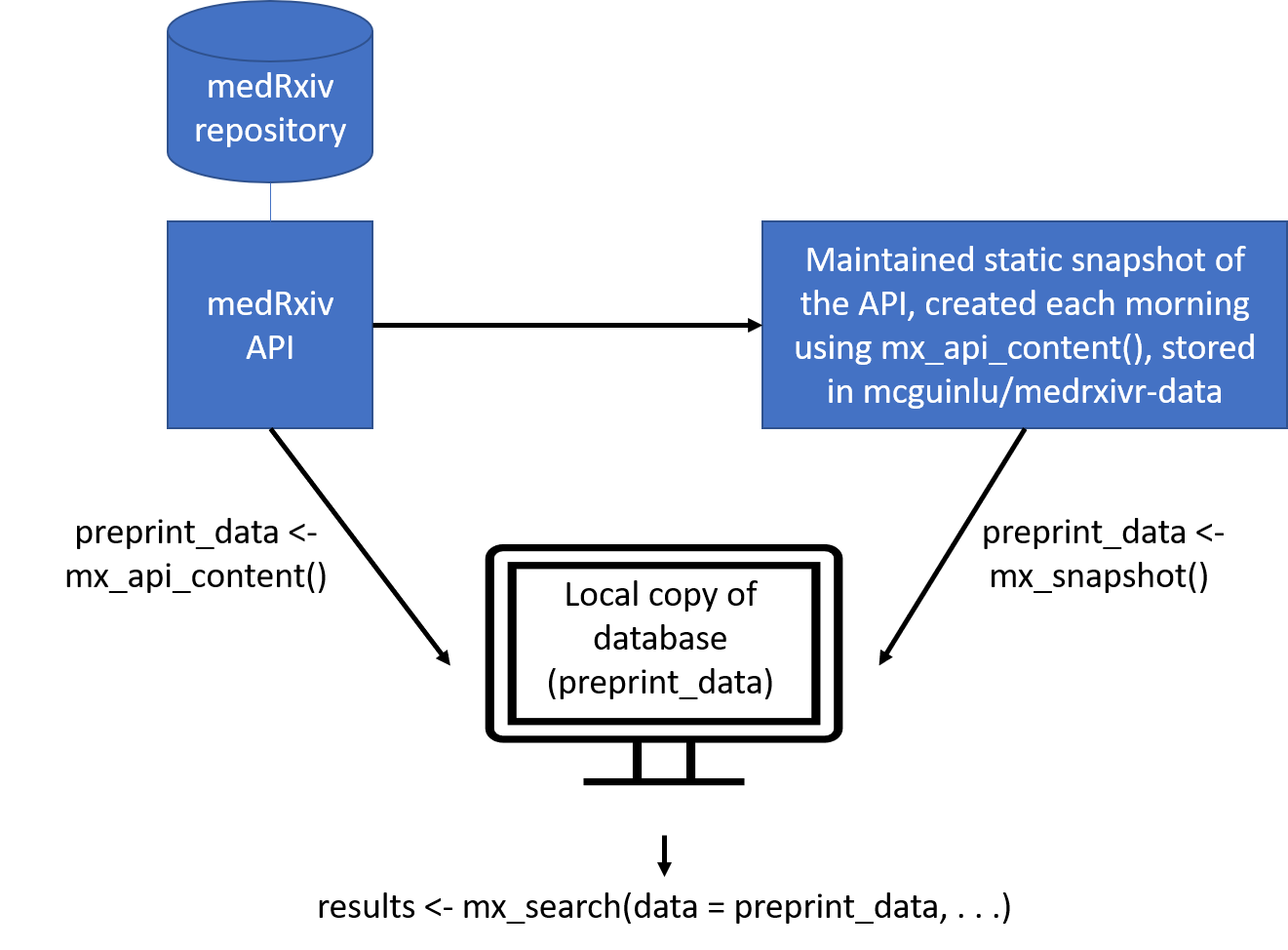
and then do custom queries, or even download article PDFs (this snippet comes from the repository README):
# Perform a simple search
results = mx_search(data = preprint_data,
query ="depression")
# Perform an advanced search
topic1 = c("dementia","vascular","alzheimer's") # Combined with Boolean OR
topic2 = c("lipids","statins","cholesterol") # Combined with Boolean OR
myquery = list(topic1, topic2) # Combined with Boolean AND
results = mx_search(data = preprint_data,
query = myquery)
Why is it useful?
When doing some kind of analysis with living, changing data, it’s important to not only be able to retrieve it programatically, but also to be able to freeze the version of the data that you use. By downloading an entire snapshot in time, this makes it easy to do your analysis over the dataset without having it change on you!
How do I cite it?
The repository DOI comes by way of Zenodo:
@article{McGuinness2020,
doi = {10.21105/joss.02651},
url = {https://doi.org/10.21105/joss.02651},
year = {2020},
publisher = {The Open Journal},
volume = {5},
number = {54},
pages = {2651},
author = {Luke A. McGuinness and Lena Schmidt},
title = {medrxivr: Accessing and searching medRxiv and bioRxiv preprint data in R},
journal = {Journal of Open Source Software}
}
How do I get started?
And if you are a developer, it would be fun to contribute new recipes for runners that are not added yet.
How do I contribute to the software survey?
or read more about annotation here. You can clone the software repository to do bulk annotation, or annotation any repository in the software database, We want annotation to be fun, straight-forward, and easy, so we will be showcasing one repository to annotate per week. If you’d like to request annotation of a particular repository (or addition to the software database) please don’t hesitate to open an issue or even a pull request.
Where can I learn more?
You might find these other resources useful:
- The Research Software Database on GitHub
- RSEpedia Documentation
- Google Docs Manuscript you are invited to contribute to.
- Annotation Documentation for RSEpedia
- Annotation Tutorial in RSEng docs
For any resource, you are encouraged to give feedback and contribute!
Recent Posts
- Posted on 21 Mar 2021
- Posted on 07 Mar 2021
- Posted on 21 Feb 2021
- Posted on 21 Feb 2021